
Multidisciplinary Neuroscientific Technologies Ltd / Harry Muzart
H: Home Page - BioNeuroTech
DCT: Developer Coding Tools, Programming Platforms, Base Code Info
RP: Research & Pre-Publication Works
A: About BNT, and Products & Services
L: Lab Experiment Methods (VR-Simu/DIY-Tech, Applications)
Computational Modelling & Implementation :
C-bCM-S: Computational (Bio)Chemical & Molecular Systems [⋆⋆]
C-NhaNp-S: Comp. NeuroHistoAnatomical & NeuroPhysiological Sys. [⋆⋆⋆⋆⋆⋆⋆]
N: Note on Neural Networks
C-OBNR-S: Comp. Organismal BioMedical & Neuro-Robotic Sys. [⋆⋆⋆⋆]
C-Cogn-S: Computational Cognitive Systems (non-A.I. & A.I.) [⋆⋆⋆⋆⋆⋆⋆]
C-mE-S: Computational Macro-Economic (Populations) Systems [⋆]
Gen: Generalist & Misc Projects [⋆⋆]
iML: Interactive Deep Machine Learning Data Apps (webpage-based) [⋆⋆]
R: Repository of External Projects
W: Website API/IDE {TBC}
D: Disclaimer (Legal Policies) / Intellectual Property / Commercial / AdSense
C-NhaNp-S: Computational NeuroHistoAnatomical & NeuroPhysiological Systems [⋆⋆⋆⋆⋆⋆⋆]
This section will cover Computational NeuroHistoAnatomical & NeuroPhysiological Systems. What is meant by 'Histo' is the Macro-Anatomy, at the level of networks of neurons. This section will cover the field of Neural Connectomics broadly.
Neuroimaging Analysis with Python (ML/non-ML)
Working on: Machine learning programming (using Python) for NeuroImaging pattern recognition, statistical comparative analytics, functional decoding, and connectomics.
Project Fields: xxxxx
Description: xxxxx
My Work & Contributions: Python programming; use on imported NeuroImaging data sets
Collaborations/Feedback: xxxxx
Location Activities: Home-based
Status/Timeline: xxxxxx
Other Links to external sources:
--- https://www.python.org/downloads/
Anaconda modules for all I-Pyhton module libraries, site packages and variable dependencies.
--- https://openfmri.org/ - open-source database dataset
--- https://www.ndcn.ox.ac.uk/divisions/fmrib/
--- http://nilearn.github.io/auto_examples/index.html
- nilearn functionalities examples
--- https://pypi.python.org/pypi/numpy - numpy imports
--- https://github.com/nilearn/nilearn
--- https://pypi.python.org/pypi/nilearn
--- http://nipy.org/packages/nilearn/index.html
--- Jupyter Notebook for prototyping code and displaying scans plots.
http://nbviewer.jupyter.org/github/ipython/ipython/blob/4.0.x/examples/IPython%20Kernel/Index.ipynb
--- http://gael-varoquaux.info/programming/nilearn-sprint-hacking-neuroimaging-machine-learning.html
--- https://www.pyimagesearch.com/2017/03/20/imagenet-vggnet-resnet-inception-xception-keras/
--- https://research.google.com/teams/brain/
--- Estève L. 2015. Big data in practice: the example of nilearn for mining brain imaging data. Scipy 2015; Jul 2015, Austin, Texas, United States. 〈hal-01207106〉
--- Halchenko Y O. 2015. Overview of statistical evaluation techniques
adopted by publicly available MVPA toolboxes . OHBM 2015, Honolulu, Hawaii, USA.
--- Smith S M, Peter T. Fox, Karla L. Miller, David C. Glahn, P. Mickle Fox, Clare E. Mackay, Nicola Filippini, Kate E. Watkins, Roberto Toro, Angela R. Laird, Christian F. Beckmanna. 2009. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A.
Hardware: Intel Quad Core i7 & Celeron Computers
Operating Programs & Software Tools: MS Win 10 OS; Anac Prmpt; Edge; JupyterNotebk; IPython CmdShell; IPython File.
Programming Languages: Python 3.6
Coding Elements: xxxxx
Computational Techniques: xxxxx
Link to Programming Code Files:
Public (full open access) (Google Drive Cloud and GitHub): https://drive.google.com/drive/folders/0B0qSKFqszohLTTZIZkg1c09xaVU?usp=sharing
Private (shared password-protected) (Google Drive Cloud): XXXXX
File types: .py; .ipynb, .png
License: xxxxx
Neuroimaging Analysis Python (ML/non-ML) (iPy/Jupy/SKL)
--- nilearn_data_dl > PNAS_Smith09_bm10.nii.gz > fMRI data (9343 KB)
--- Nilearn1.ipynb (1484 KB)
--- Nilearn1-22oct2017-1b.ipynb (1443 KB)
--- hm-mod-plt_canica_resting_state.ipynb (408 KB)
--- hm-mod-plot_classifier_comparison.py (5 KB)
--- hm-mod-plot_localizer_simple_analysis.ipynb (36 KB)
--- hm-mod-plot_oasis_vbm_space_net.ipynb (90 KB)
--- hm-mod-plot_prob_atlas.ipynb (1109 KB)
--- nilearndecod-8343&#g80394(hmuz)_ipynb.docx
Next step after all this:
--- Computer vision on digitally printed histology scans of the hippocampal BNNs; visual system ANNs for pattern recognition of hippocampal region activity. The other examples would be straight to computational models of the hippocampus, frontal cortex, amygdala. Much faster and more efficient detection of clinical abnormality in brain vs normal.
--- Make an Android App (using AStud 3.0 with JDK 8.0, programmed with OO/class-based Java PL & xml layouts for front-end) (and C++ for native web-based back-end with Google Cloud Platform & TensorFlow ImageNets) (use of mobile device camera) (and open-source collab GitHub for faster debugging & learning & testing) (will also need access to NeuroImaging dataset file repository)
Base Math Coding & Program Computations & System (Diagrammatic):
XXXXX
Images:
Content Files Edits by HM. Content: Dataset Images and Programming Env Platforms by others. All Images are Screenshots by HM.
Video Embeds: xxxxx




.png)

Slicer3D (fMRI/DTT data)
xxxxxx
xxxxx
Structural & Functional Magnetic Resonance Imaging and Diffusion Tensor Tractography
LINKS:
https://www.slicer.org/wiki/Documentation/Nightly/Developers
https://github.com/Slicer
http://www.spl.harvard.edu/publications/pages/display/?collection=11
https://www.python.org/downloads/source/
https://openfmri.org/dataset/
https://nifti.nimh.nih.gov/
https://discourse.slicer.org/u/Harry-Muzart/summary
https://www.bioneurotech.com
https://github.com/Harry-Muzart
Developer tools for processing nii/nifti files, see:
https://www.natbrainlab.co.uk/atlas-maps
https://www.facebase.org/help/viewing-nifti/
http://people.cas.sc.edu/rorden/mricron/index.html
http://ric.uthscsa.edu/mango/develop.html
https://imagej.nih.gov/ij/developer/index.html
http://mbat.loni.usc.edu/plugins/
https://www.mccauslandcenter.sc.edu/crnl/
https://www.nitrc.org/plugins/mwiki/index.php/surfice:MainPage
https://www.mccauslandcenter.sc.edu/mricrogl/source
https://www.mccauslandcenter.sc.edu/crnl/c-star
fMRI SPM/Matlab Neuroimaging - Statistical Parametric Mapping
(2014-2018) (2019-2020) (2020-2021)
Analyses and manipulations by H.M. - to test novel hypotheses with existing data - currently in progress ....
see original software, data-sets and documentation available at:
https://www.fil.ion.ucl.ac.uk/spm/
Penny, Friston, Ashburner, et al., (2011) "Statistical Parametric Mapping"
Matlab
https://uk.mathworks.com/products.html?s_tid=gn_ps
 | 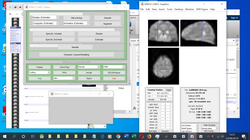 |
|---|---|
 |  |
 |  |
 |  |
Neural Networks (SimBrain 3.0)
● Other projects I have worked on involves Modelling Biological Neural Networks and Artificial Neural Networks. This type of Neural Engineering has huge applications.
► SIMBRAIN (File modification by H Muzart):
• Project Fields: xxxxx
• Description: xxxxx
• My Work & Contributions: User-Interface level NN manipulations, and base code reprogramming.
• Collaborations/Feedback: xxxxx
• Location Activities: Home-based
• Status/Timeline: xxxxxx
Other Links to external sources:
--- http://www.simbrain.net/Downloads/downloads_main.html
--- http://simbrain.net/Documentation/apidocs/index.html
--- https://github.com/simbrain/simbrain
--- http://www.sciencedirect.com/science/article/pii/S0893608016300879
--- Tosi Z, Yoshimi J (2016) “Simbrain 3.0: A flexible, visually-oriented neural network simulator”. Journal of Neural Networks. PMID: 27541049. doi: https://doi.org/10.1016/j.neunet.2016...
Zachary Tosi, Jeffrey Yoshimi.
Hardware: Intel Quad Core i7 & Celeron Computers
Operating Programs & Software Tools: MS Win 10 OS; Java Env; SimBrain 3.02
Programming Languages: xxxxxx
Coding Elements: xxxxxx
Computational Techniques: xxxxxxxxxx
Link to Programming Code Files:
Public (full open access) (Google Drive Cloud and GitHub): https://drive.google.com/drive/folders/0B0qSKFqszohLTTZIZkg1c09xaVU?usp=sharing
Private (shared password-protected) (Google Drive Cloud): XXXXX
File types: xxxxxx
License: xxxxxx
--- workspacehm1 (folder of files) (605 KB)
--- workspace_hm3e.zip (1 file) (5KB)
Base Math Coding & Program Computations & System (Diagrammatic):
Xxxxxx
Images:
Content Files Edits by HM, Screenshots by HM.
Video:
You can see that if the input pattern of neural activations (set of initial potential values in the multi-parametric network) changes – the overall value of the network still converges towards a common point (attractor network). This is because the neurons and connections have not been modified. When the system is changed (ie. synaptic connection weights are modified (therefore affecting the internodal learning algorithms) or when neurons degenerate & are lost in disease), then the local mimimum in the search space landscape changes (new attractor point).
I am interested in modelling various systems in the brain:
--- MTL/Hippocampal/CA3 neural networks. Hopfield recurrent networks, attractor network, with pattern converging towards a common pattern (memory), based on initial cues. Auto-associative networks in CA3, content-addressable (not location-addressable) memory – re-construction of whole memory from one or more cues. Sparse code and system of overlaps – memories shared info (are associated) but non-spurious distinct memories can be formed from the system. Pattern separation and auto-association and pattern completion processes. Unsupervised self-organising topographic maps, STDP, rate coding, and oscillatory interference, for representation of allocentric space, context-based episodic memory, predictive navigation of scenes.
--- V1-V4/V5 system of deep convolutional neural networks for visual feature representations.
--- Limbic/Amygdala/OFC system for reward-based and aversion-based semi-supervised reinforcement learning.
--- The PFC neural networks for top-down conscious decision-making.
--- Somatosensory and somatomotor systems.
see https://www.bioneurotech.com
 | 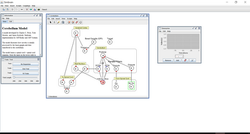 |  |
|---|---|---|
 |
Histology Samples (demos by HM)
 |  |
|---|---|
 |  |
 |
Neural Nets (NeuroPh / Java)
Other Links to external sources:
http://neuroph.sourceforge.net/features.html
https://netbeans.org/community/index.html
https://opensource.org/docs/osd
http://www.apache.org/licenses/LICENSE-2.0.html
http://neuroph.sourceforge.net/developer_guidelines.html
https://github.com/neuroph/neuroph/tree/master/neuroph-2.9
--- NewDataSet1.tset 1 KB NetBeans Training Set
--- neuralTest_hm4 1KB Test File Data Columns

Google's Tensorflow Playground NN - repurposing in JvS/TypeScript
https://github.com/Harry-Muzart/tensorflow
Other Links to external sources:
http://playground.tensorflow.org
https://github.com/tensorflow/playground
https://www.tensorflow.org/api_docs/serving/
http://scikit-learn.org/stable/modules/decomposition.html#decompositions
Model visualization and coding deep NN layers
http://tflearn.org/layers/conv/
https://www.google.com/policies/terms/
https://www.apache.org/licenses/LICENSE-2.0
My files:
https://drive.google.com/drive/folders/0B0qSKFqszohLTTZIZkg1c09xaVU?usp=sharing
--- seedrandom.d.ts (1KB)
--- nn_hm7.ts (13KB)
Images:
Content File Manips/Edits by Me, Screenshots by Me.
 |  |  |
|---|
Graphical Representations (3D Matrices) (SciLab 6.0)
Project Fields: xxxxx
Description: xxxxx
My Work & Contributions: xxxxx
Collaborations/Feedback: xxxxx
Location Activities: xxxxx
Status/Timeline: xxxxxx
Other Links to external sources:
Free open source:
--- https://www.scilab.org/projects/open_source_involvement
--- https://scilab.io/cloud/web-application/
--- https://www.inria.fr/en/teams/scilab
--- https://jamesmccaffrey.wordpress.com/2011/05/13/creating-a-3d-plot-with-scilab/
--- http://www.matrixlab-examples.com/scilab-3d-plot.html
--- GNU octave syntax https://www.gnu.org/software/octave/
Fee-based:
--- MatLab https://www.mathworks.com/solutions.html?s_tid=gn_sol
--- Maple https://www.maplesoft.com/products/Maple/
Hardware: Intel Quad Core i7 & Celeron Computers
Operating Programs & Software Tools: MS Win 10 OS
Programming Languages: SciLab Prog Lang
Coding Elements: mesh coords, f(x)àx,z
Computational Techniques: xxxxx
Link to Programming Code Files:
Public (full open access) (Google Drive Cloud and GitHub): https://drive.google.com/drive/folders/0B0qSKFqszohLTTZIZkg1c09xaVU?usp=sharing
Private (shared password-protected) (Google Drive Cloud): XXXXX
File types: .sce
License: xxxxx
--- octave1 (1 KB)
--- scilan1hm.sce (2KB)
Base Math Coding & Program Computations & System (Diagrammatic):
XXXXX
Images:
Content Files by HM, Screenshots by HM.
Video Embeds: xxxxx
 |  |
|---|---|
 |
Neural Networks (Emergent PDP++)
Project Fields: xxxxx
Description: xxxxx
My Work & Contributions: xxxxx
Collaborations/Feedback: xxxxx
Location Activities: Home-based
Status/Timeline: xxxxxx
Other Links to external sources:
--- https://grey.colorado.edu/emergent/index.php/Main_Page
--- https://www.ncbi.nlm.nih.gov/pubmed/18684591
--- https://senselab.med.yale.edu/MicrocircuitDB/ModelList.cshtml?id=53430
--- http://ski.clps.brown.edu/cogsim/Emergent_v5.3.2_manual.pdf
--- https://www.researchgate.net/publication/23153539_The_Emergent_Neural_Modeling_System
--- http://neurobot.bio.auth.gr/2007/emergent-neural-network-simulation-software-formerly-pdp/
--- http://www.brains-minds-media.org/archive/1406
--- Emergent PDP++ LeabRa: The Artificial Neural Networks Algorithm Leabra
https://cran.r-project.org/web/packages/leabRa/
Hardware: Intel Quad Core i7 & Celeron Computers
Operating Programs & Software Tools: MS Win 10 OS; xxxxx
Programming Languages: xxxxx
Coding Elements: xxxxx
Computational Techniques: xxxxxxx
Link to Programming Code Files:
Public (full open access) (Google Drive Cloud and GitHub): https://drive.google.com/drive/folders/0B0qSKFqszohLTTZIZkg1c09xaVU?usp=sharing
Private (shared password-protected) (Google Drive Cloud): XXXXX
File types: xxxxx
License: xxxxx
--- XXXXXXXX
Base Math Coding & Program Computations & System (Diagrammatic):
XXXXX
Images:
Content Files by Me, Screenshots by Me.
Video Embeds: xxxxx
 |
|---|
Object3D (Blender3D) (with Python 3.6 functionalities)
Project Fields: xxxxx
Description: xxxxx
My Work & Contributions: xxxxx
Collaborations/Feedback: xxxxx
Location Activities: xxxxx
Status/Timeline: xxxxxx
Other Links to external sources:
--- https://docs.blender.org/manual/en/dev/editors/python_console.html
--- https://www.blender.org/foundation/development-fund/
--- https://www.blender.org/foundation/donation-payment/
Hardware: Intel Quad Core i7 & Celeron Computers
Operating Programs & Software Tools: MS Win 10 OS; xxxxxxx
Programming Languages: xxxxxx
Coding Elements: xxxxxx
Computational Techniques: xxxxxx
Link to Programming Code Files:
Public (full open access) (Google Drive Cloud and GitHub): https://drive.google.com/drive/folders/0B0qSKFqszohLTTZIZkg1c09xaVU?usp=sharing
Private (shared password-protected) (Google Drive Cloud): XXXXX
File types: xxxxxx
License: xxxxxx
--- NN_hm1.blend (X KB) (Priv)
Base Math Coding & Program Computations & System (Diagrammatic):
XXXXX
Images:
Content Files by Me, Screenshots by Me
Video Embeds: xxxxx
 |  |
|---|---|
 |
Human Connectome Projects, Hippocampal Neural Network Architecture
See my literature review posts at:
HM HumanBrainProject - https://forum.humanbrainproject.eu/users/harry_muzart/activity
Meta-analysis upcoming
e.g. PFC-Limbic/Hippocampal circuitry models in one-shot learning, imagination, predictive models, policy & value networks for creativity, reward-based reinforcement learning, unsupervised episodic memory, long short term memory, content addressable spatio-temporal auto-association and episodic memory models, hierarchical deep structuring, spatial navigational dynamics, comparative indexing from deep convolutional neural nets.
#########################################################
EEG Oscil and ERP Data / Statistical R Programming / Brainstorm (Matlab/Java)
Project Fields: xxxxxx
Description: xxxxxx
My Work & Contributions: xxxxxx.
Collaborations/Feedback: xxxxxx
Location Activities: xxxxxx
Status/Timeline: xxxxxx
Other Links to external sources:
--- http://developer.r-project.org/
--- https://cran.r-project.org/mirrors.html
--- http://www.ucl.ac.uk/lifelearning/courses/statistical-computing-r-programming-introduction
--- https://engineuring.wordpress.com/2009/07/08/downloadable-eeg-data/
--- Data Files
https://cran.r-project.org/web/packages/eegkit/eegkit.pdf
Hardware: Intel Quad Core i7 & Celeron Computers;
Operating Programs & Software Tools: MS Win 10 OS; xxxxxxxxxx
Programming Languages: R
Coding Elements: xxxxx
Computational Techniques: xxxxxxxxxx
Link to Programming Code Files:
Public (full open access) (Google Drive Cloud and GitHub): https://drive.google.com/drive/folders/0B0qSKFqszohLTTZIZkg1c09xaVU?usp=sharing
Private (shared password-protected) (Google Drive Cloud): XXXXX
File types: xxxxx
License: xxxxx
--- XXXXXX
Base Math Coding & Program Computations & System (Diagrammatic):
XXXXX
Images:
Content Files by Me, Screenshots by Me
Video Embeds: xxxxx
#########################################################
EEG-based DIY Brain-Machine Interfaces (non-invasive) (mobile/portable/wireless)
see https://www.bioneurotech.com/obmnr
Brain Mapping (using Allen Explorer 2.0)
Project Fields: xxxxx
Description: xxxxx
My Work & Contributions: xxxxx
Collaborations/Feedback: xxxxx
Location Activities: xxxxx
Status/Timeline: xxxxxx
Other Links to external sources:
http://human.brain-map.org/static/brainexplorer
http://www.nvidia.com/Download/index.aspx?lang=en-us
http://alleninstitute.org/legal/terms-use/
Hardware: Intel Quad Core i7 & Celeron Computers;
Operating Programs & Software Tools: MS Win 10 OS; xxxxxxxxxx
Programming Languages: xxxxx
Coding Elements: xxxxx
Computational Techniques: xxxxxxxxxx
Link to Programming Code Files:
Public (full open access) (Google Drive Cloud and GitHub): https://drive.google.com/drive/folders/0B0qSKFqszohLTTZIZkg1c09xaVU?usp=sharing
Private (shared password-protected) (Google Drive Cloud): XXXXX
File types: xxxxx
License: xxxxx
--- XXXXXX
Base Math Coding & Program Computations & System (Diagrammatic):
XXXXX
Images:
Content Manipulations by Me, Screenshots by Me
Video Embeds: xxxxx
 |
|---|
Other
Other future works will be related to these other external independent projects by other people/organisations listed below.
Here is a new list :
► https://sites.google.com/site/bioneurotech/-repo-2
► https://docs.google.com/document/d/1Nv6BimGLFgvbSYeNQZO5Cjna_iWcquwoF47MMsll5fA/edit#
#### My Works/Projects status (being researched or in progress/pre-completion) ####
(I will create my own programs using these () programs )
--- NN arrays, Machine Learning (Python using numpy, scipy, matplotlib, scikit-learn; tensorflow, Inception, MNIST; Python IDE & exe prompt, Docker Virtual Box Manager)
--- Java JDK, JSE, JRE, 7.0-8.0
--- Artificial Neural Networks
--- Long Short-Term Memory
--- Deep Convolutional neural networks
--- Windows 3D
--- NEURON – simulation environment for individual neuron electrophysiological dynamics. Also standard compartmental models of neurons and networks of neurons
--- ERP/EEG
--- NeuGen 1.7
--- Emergent PDP++ - neural simulation software (Munakata Comp Expl in Cogn NEur)
--- Neuroph
--- ModeDB
--- SciLab 6.0 (Trappenberg – Fund Comp Neuro)
--- Octave 4.2 (Trappenberg) (indirectly contributes to open source science community using octave)
--- Weka 8.8 (Witten – Data mining) [Installed]
--- SpikeFun from DigitalCortex
--- Allen Brain Explorer 2
--- R Statistical programming
--- MinGW C/C++ Programs
--- Tensorflow Playground experiments
--- Tensorflow Playground (by Google) repurposing from Github and other Tensorflow repositories projects (Python; TypeScript, Haskell, HTML, C++, Scala; Jupyter Notebook)
--- MatLab (see KCL group for neuro robotics matlab code)
--- Google Cloud TPUs
--- Neuroscience Gateway NSG – run (for free) parallel simulations on supercomputers
--- Genesis 2.5/3
--- PyGenesis
--- inSilico (C++ for computational neuroscience)
--- MOOSE (Python, XML, NeuroML)
--- NeuroML
--- NEST
--- Brian – Spiking neural networks
--- CARLsim – GPU-acc API for synaptic and neu network level – automated parameter tuning by evolutionary algos.
--- NEurokernel (Python & CUDA)
--- NEOSIM2 – (parallel kernel for spiking neurons, code with NEuroML, visualise with Java2D)
--- PRonTo (UCL)
--- neuroConstruct (UCL) (SWX, Neurolucida, ChannelML, MorphML, Python)
--- Neurolucida Explorer (microscopic morphometry)
--- StdpC/DynClamp2
--- FreeSurfer for neuroimaging
--- Helmholtz for modular database management in neuroscience (Python, Django, MySQL, SQLite, Oracle web frameworks)
--- NeuralEnsemble (incl PyNN, OpenElectrophy, libNeuroML)
--- BluePyOpt
--- NetPyNE
--- Virtual Brain Personalised Multimodal Pipeline
--- CONICAL (C++)
--- Slide Surfer Histology (used previously at UCL)
--- MatLab
--- Maple
--- Open Vibe, Open CBICIE
--- Hopfieldsimulator.codeplex.com
--- github > retina
--- NeuroQuantite – for development of neural network axons and dendrites
--- 3D Slicer (neuroimaging) (also neuroimaging of brain sub-areas, neural layers an columns: Amira 3D, AFNI, Analyze (BIR MC), BIAP, BrainSuite, CamBA, CARET, CONN< EEGLAB, EXploreDTI, FMRIB SL, ISAS, LONI, Mango, NP Biomkarker Toolbox, NITRC, nordicICE, SDM, SCT, SPM, TORTOISE).
--- C++ ANNs: OPenNN, Qt Cretor (pay), CMake, Visual C++,
Tensorflow C++ API.
--- Allen Cell Explorer
#### My future direct Contributions to larger/wider (even more complex) External Projects/Platforms by others ####
--- Github Tensorflow
--- G DeepMind Health PPI/Implem/Apps, etc
Plan for future involvement in Future:
--- Human Brain Project & Human Connectome Project Forum & GitHub Collaboration
--- HBP (EU) (will join soon)
--- Human Connectome
--- Robotics (BostonDynamics, Hanson, etc)
--- AI
--- Google Verily, Google X
--- Brain Machine Interface in Clinical Neurology
--- Materials and Tissues
--- NeuroLink, NeuroLace Technology
--- Neuro-Endo/Exoskeleton (see Elon Musk investment portfolio) (similar opensource is xxxxx – will request to contribute in 2022)
My other current Experimental Work:
--- For all current files, see link:
https://drive.google.com/drive/folders/0B0qSKFqszohLTTZIZkg1c09xaVU?usp=sharing
--- SimBrain Experiments
--- Allen BioArray Data Comparatives
--- TensorFlow NN repurposing in GitHub
--- ERP / EEG data analysis
--- Comparative Data in Excel VBA / CSV / Access Files - Data between Hippocampus and PFC can generated;
--- To be confirmed:Use of Machine Learning Analytics on the above Data Projects
####################
Intelligent Analytics
--- Artificial Intelligence, Deep Machine Learning, Cognitive Statistical Analytics, Sentiment Analysis, and Their Applications. {Intelligent Analytics}
### ML with Python and Google Cloud
### Databases projects
### App Script Chrome Ext
### Scratch up own web domain linkings
--- Further Applicaitons
### Applications of Intelligent Systems --- GDMH
I am also interested in other projects:
External Projects:
Solving the code for Emotional Episodic Memories - just one cognitive trait of humans. (based on my BSc research at UCL with staff from the NPP Dept and ICN) - see diagrams above.
Work inspired by Oxbridge, KCL, UCL & Imperial (London, UK) researchers: e.g. Demis Hassabis, Neil Burgess, Peter Dayan, Geraint Rees, Eleanor Maguire, John O'Keefe, Karl Friston, Ray Dolan, Matteo Carandini, Thore Graepel, and many others.
Interesting:
see https://www.researchgate.net/project/A-computational-framework-for-the-embodied-mind for neural circuit cognitive implementation
see https://www.academia.edu/Documents/in/Robotics_and_AI for neural circuit behavioral implementation in humanoid robots
Other projects I am currently looking into & researching, as stated above:
---1--- Machine Learning Pattern Recognition
---2--- Use of MLPR on: High-resolution 3D fMRI Scans (Cortical & Sub-cortical Biological Neural Networks)
---3--- For practical applications in Brain-Machine Interface Physiology and Clinical conditions.
Some external links:
Residual and Plain Convolutional Neural Networks for 3D Brain MRI Classification (Korlev et al 2017)
https://arxiv.org/pdf/1701.06643
PRoNTo Software - MLNL - UCL
http://www.mlnl.cs.ucl.ac.uk/pronto/prtsoftware.html
Nilearn: Machine learning for NeuroImaging in Python
nilearn.github.io/
Tom Mitchell's fMRI Research Group Page
move this to Organismal Biomedi
Google DeepMind & #DMHealth:
https://deepmind.com/research/publications/
https://deepmind.com/research/alphago/
https://deepmind.com/research/open-source/
https://deepmind.com/applied/deepmind-health/
https://deepmind.com/applied/deepmind-health/working-nhs/how-were-helping-today/
My GitHub:
https://github.com/Harry-Muzart
Chu et al 2016 - ML, MRI, tumour radiotherapy
https://storage.googleapis.com/dmhir-documents/UCLH_radiotherapy_protocol.pdf
https://f1000research.com/articles/5-2104/v1
Information Processing in Multi-scale Models of Neurons and Neural Networks
http://www.kurzweilai.net/a-computer-like-brain-mechanism-that-makes-sense-of-novel-situations
Simulation of Spiking Thalamocortical Brain Network 3 - 3D View, 1M Neurons, 189M Synapses
https://www.youtube.com/watch?v=PXTLXXdDVMM
Allen Brain Atalas Connectomics
https://alleninstitute.org/about/
http://human.brain-map.org/static/brainexplorer
http://mouse.brain-map.org/static/atlas
(Images below: all screenshots by me, BNT/HFM)




---

---
https://commons.wikimedia.org; Rotating_brain_colored.gif
https://commons.wikimedia.org; Isat.gif
#




